TLDR: It is pretty cool that you can directly leverage Deepmind’s cutting edge AI powered AlphaFold and AlphaMissense projects to get insight into your own genome!
Google DeepMind created AlphaMissense, new AI model that predicts the pathogenic classification of the effects of ‘missense’ mutations. Using this tool they created and released a catalogue of ‘missense’ mutations where researchers can learn more about what effect they may have.
In a paper published in Science, DeepMind show it categorised 89% of all 71 million possible missense variants as either likely pathogenic or likely benign. By contrast, only 0.1% have been confirmed by human experts.
Missense variants are genetic mutations that can affect the function of human proteins. In some cases, they can lead to diseases such as cystic fibrosis, sickle-cell anaemia, or cancer.
AlphaMissense builds on DeepMind’s AlphaFold protein folding prediction project. AlphaFold predicts the 3D structures of proteins based on their amino acid sequences. AlphaMissense leverages structural insights into amino acid sequences provided by AlphaFold to classify variants as benign, ambiguous or pathogenic based on potential health impacts.
Read the DeepMindblog post and their published paper.
Visualization from the DeepMind blog post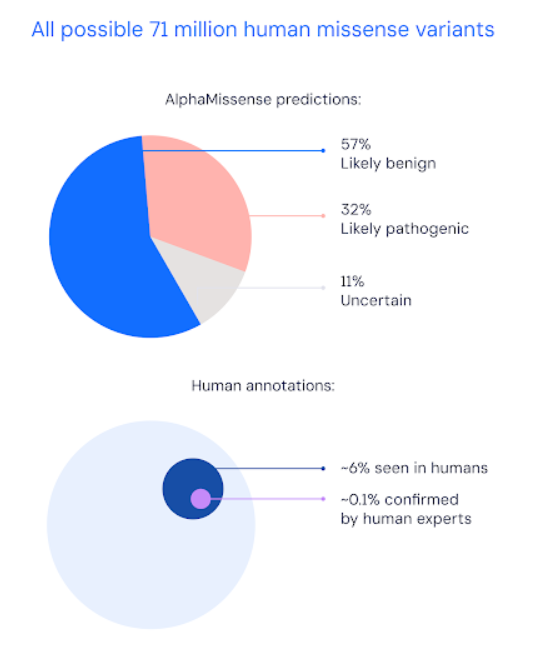
Matching 23andMe genome data with AlphaMissense data => benign, ambiguous or pathogenic
A practical use of the AlphaMissense missense variants is matching it to your 23andMe genome data. Both sets of data have a Reference SNP cluster ID (rsID) value which looks something like “rs2817580”. rsIDs are unique identifiers assigned to specific genetic variations (like single nucleotide polymorphisms, or SNPs) which are maintained in the US National Library of Medicine dbSNP database.
A script Github link developed by developer Edouard Belval and shared in a Hacker News post matchs your 23andMe genome data to the AlphaMissense missense data and prints out mutation percentages along with their classification as benign, ambiguous or pathogenic, in addition to a nice visualization of variations by allele count.
The script will give you a summary of the matching results:
% Mutated genes: 1.97%
% Mutated genes with at least 1 allele classified as benign: 93.98%
% Mutated genes with at least 1 allele classified as ambiguous: 3.94%
% Mutated genes with at least 1 allele classified as pathogenic: 2.08%
% Mutated genes with 2 alleles classified as benign: 31.11%
% Mutated genes with 2 alleles classified as ambiguous: 0.88%
% Mutated genes with 2 alleles classified as pathogenic: 0.33%
You can also output a list of your 23andMe genome data that had rsIDs that could be matched (~2,000 records) to see individual snp’s AlphaMissense missense pathengenicity value and classification as benign, ambiguous or pathogenic:
rs3748597 0.0564 benign
rs1059831 0.3098 benign
rs7535528 0.0892 benign
rs3748816 0.0458 benign
i6059967 0.0458 benign
rs2185639 0.0729 benign
If you use 23andMe, the genome data it is relatively simple to download. You just need to export your 23andMe data which will include a genome.txt file. Then you can reference this file in Belval ‘s Github repo code to match the AlphaMissense missense data to your 23andMe genome data and get outputs simialar those above.
After matching 23andMe genome data with AlphaMissense data
After doing the matching I recommend doing some manual searches of your pathogenic rsID’s against the dbSNP database which works like a search engine. “Pathogenic” sounds pretty dire but the variant clinical signifance might be very low or irrelevant. For example, variants might manifest only in embryonic development or have potential physical manifestation that is not evident for you.
Searching is easy. You simply search by the rsID to find the rsID documentation which includes descriptions of clinical significance, publications, history, detailed descriptions of location in genome, and more.
Next steps
Automating the retrieval of dbSNP database clinical significance would be a good next project for someone. It could use LLM RAG targeted search against the dbSNP database to retrieve text summarizing what is known for the rsID. Perhaps something like sentiment analysis of the text content.
More reading
Hacker News post
Edouard Belval ‘s Github repo code to match the AlphaMissense missense data to your 23andMe genome data and get outputs simialar those above
DeepMind AlphaMissense blog post
DeepMind AlphaMissense published paper
DeepMind’s AlphaFold
The Illustrated AlphaFold – A visual walkthrough of the AlphaFold3 architecture, with more details and diagrams than you were probably looking for. https://elanapearl.github.io/blog/2024/the-illustrated-alphafold